Abstract
Procoagulant platelets, a subpopulation of platelets which support thrombin generation, play a role in regulating pathological thrombosis. Using a novel flow cytometry assay (Hua et at, Blood 2015) to identify procoagulant platelets based upon the cell death marker GSAO and activation marker P-selectin, we found that patients with angiogram proven coronary artery disease had heightened thrombin procoagulant platelet formation compared to healthy controls, (25.2% vs 12.2%, p<0.001). Inhibition of this is an attractive target for atherothrombosis prevention. However, the mechanism of thrombin induced procoagulant platelet formation remains controversial with discrepant results regarding requirement for the thrombin binding domain of N terminal GPIbα (Dorman et al, Blood 2000 and Ravanat et al, Blood 2010) and recent reports implicating PAR4 (French et al JTH, 2016). Here we use a recently described novel assay to investigate the roles of PARs and GPIbα measuring procoagulant platelets by FACS (GSAO+/P-selectin+) in whole blood, plasma and washed platelets after treatment with heparin, PAR and GPIbα inhibitors prior to thrombin or PAR agonist stimulation.
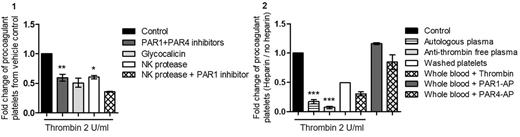
First, using active site inhibited PPACK-thrombin we demonstrated that catalytically active thrombin is required for procoagulant platelet formation in our in vitro system. PAR1 and PAR4 are implicated in procoagulant platelet formation, however, preincubation with combination of PAR1 and PAR 4 inhibitors (vorapaxar and a PAR4 polyclonal inhibitory antibody), resulted in only partial inhibition of thrombin induced procoagulant platelets (reduction 40%±5.7%, p<0.01) suggesting a role for an additional non-PAR pathway. Thrombin-GPIbα interaction was implied when competition studies with recombinant soluble GPIbα, glycocalicin showed a 49%±7.7% reduction. Involvement of the N-terminus of GPIbα was confirmed by a 39.5%±2.9% reduction after cleavage of GPIbα proximal to the thrombin binding site by NK protease. Pretreatment with PAR1 inhibitor after NK protease cleavage of GPIbα reduced thrombin stimulated procoagulant platelet formation to unstimulated levels implying both PAR cleavage and GPIbα-thrombin interaction is required for thrombin induced procoagulant platelet formation.
Consistent with a role of thrombin-GPIbα interaction, we noted a reduction in thrombin induced procoagulant platelets after heparin bolus in patients undergoing coronary angiogram (16.3±10.5% vs 6.1%±2.4%, p<0.001). Heparin is known to known to displace thrombin from GPIbα in addition to anti-thrombin effects via enhancement of antithrombin (AT). Using platelets resuspended in AT-deficient plasma, we demonstrated high dose heparin suppressed procoagulant platelet formation via an AT-independent mechanism, this was confirmed by heparin suppression of thrombin induced procoagulant platelets in washed platelets. In similar experiments, the heparinoid danaparoid, which has no anti-thrombin effect, also inhibited thrombin induced procoagulant platelet formation. Heparin inhibition was not due to PAR inhibition as heparin had no effect on direct PAR activation by PAR1 agonist TRAP or PAR4 agonist AYPGKF. These data suggest heparin has a direct effect on platelets in suppression of thrombin induced procoagulant platelet formation, most likely relating to displacement of thrombin from GPIbα.
Together this data indicates that a thrombin-GPIbα interaction via the N terminal fragment is required in addition to thrombin mediated PAR activation. This mechanism is a potential target for modifying atherothrombotic risk.
Figure 1: Whole blood was treated with PAR1 and PAR4 inhibitors and washed platelets were treated with glycocalicin, NK protease or NK and PAR1 inhibitor prior to thrombin stimulation (2 U/mL). Data are expressed as fold change from vehicle control ((*p<0.05; **p<0.01, statistics are calculated for n≥3). Figure 2: Plasma, washed platelets or whole blood were treated or not with heparin (12.8 U/ml) prior to thrombin stimulation (2 U/mL), PAR1 (20 µM) or PAR4 (300 µM) agonists. Data are expressed as the ratio of heparin-treated sample to untreated (***p<0.001, statistics are calculated for n≥3).
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal